OHBM 2023 poster supplimentary results
This year I will be attending the Organization for Human Brain Mapping’s (OHBM) 2023 conference in Montreal. I will be presenting a poster titled “Classifying Alzheimer’s disease and minor cognitive impairment using thalamic segmentation methods” My poster number is 854, and I will be available at my poster at the following times:
- Sunday, July 23, 2023: 12:15 PM – 02:15 PM (EDT)
- Monday, July 24, 2023: 01:00 PM – 03:00 PM (EDT)
If you would like to speak to me about my poster but I am not currently here, please reach out to me via twitter @neuroBren!
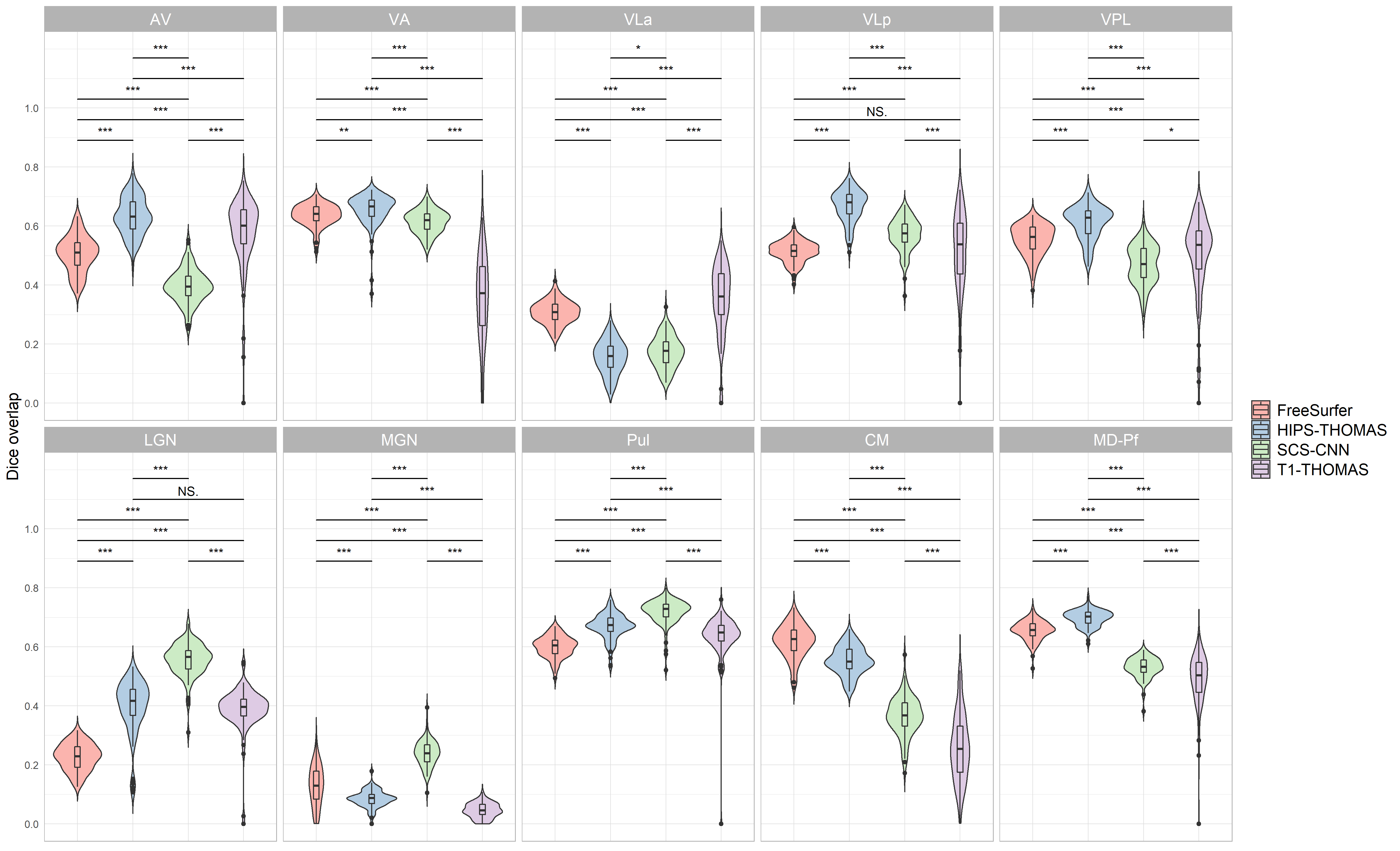
Supplementary figure 1:
Violin plots of Dice overlap between right hemisphere nuclei segmented from Human Connectome Project data using FreeSurfer, HIPS THOMAS, CNN SCS, and T1 THOMAS approaches. Two way ANOVAs found significant main effects of segmentation approach and hemisphere for all nuclei (except for VPL, which a non significant main effect of hemisphere), and a significant interaction between segmentation approach and hemisphere for all nuclei. Posthoc t test results (Bonferroni corrected) are presented to show pairwise difference between segmentation approaches for each nucleus (*p<0.05, **p<0.01, ***p<0.001).
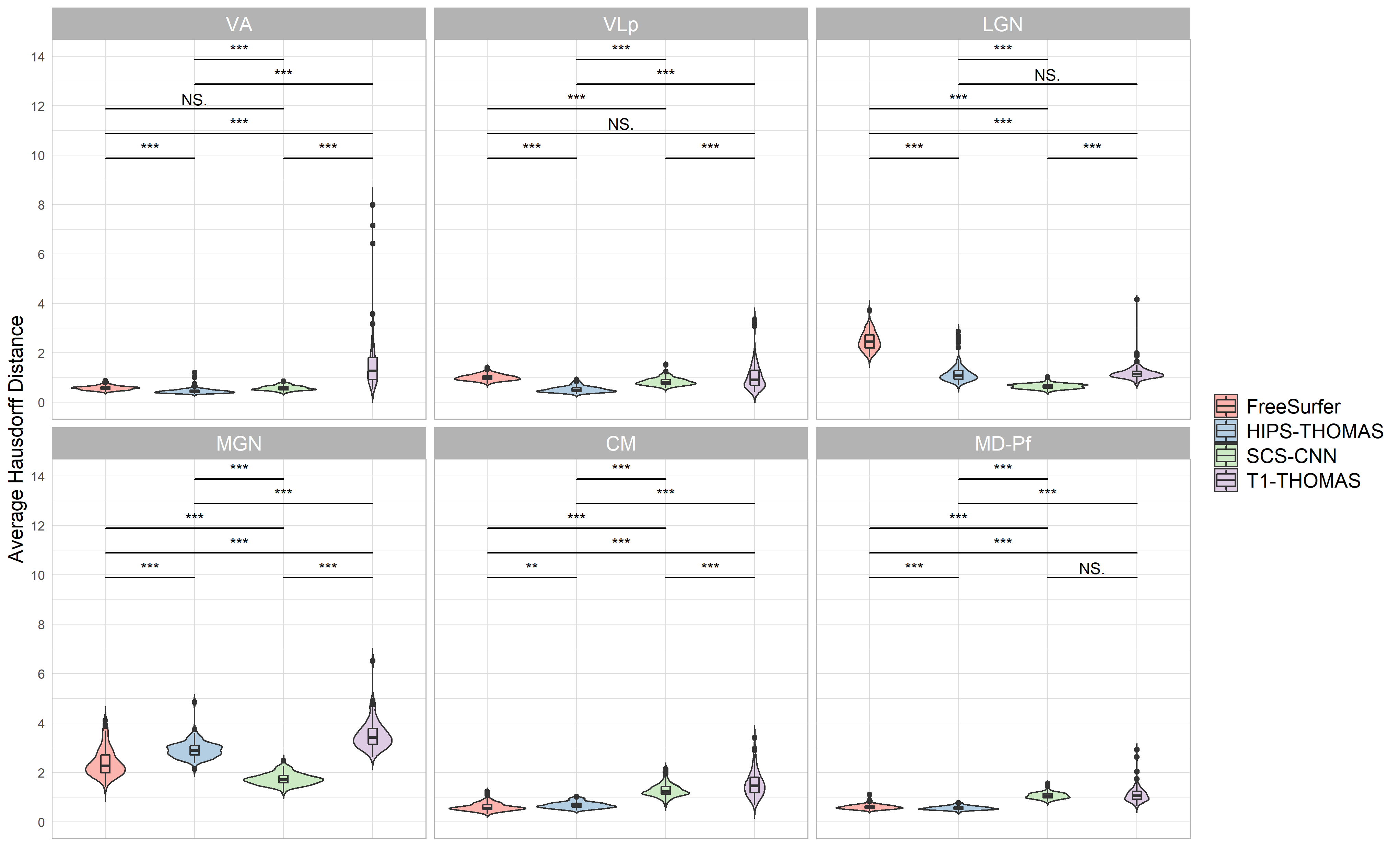
Supplementary figure 2:
Violin plots of Average Hausdorff Distance for right hemisphere nuclei segmented from Human Connectome Project data using FreeSurfer, HIPS THOMAS, CNN SCS, and T1 THOMAS approaches. Significant main effects of segmentation approach were found for VA, VLp, LGN, MGN, CM, and Md Pf, and posthoc t test results (Bonferroni corrected) are presented in the bottom plot (**p<0.01, ***p<0.001).

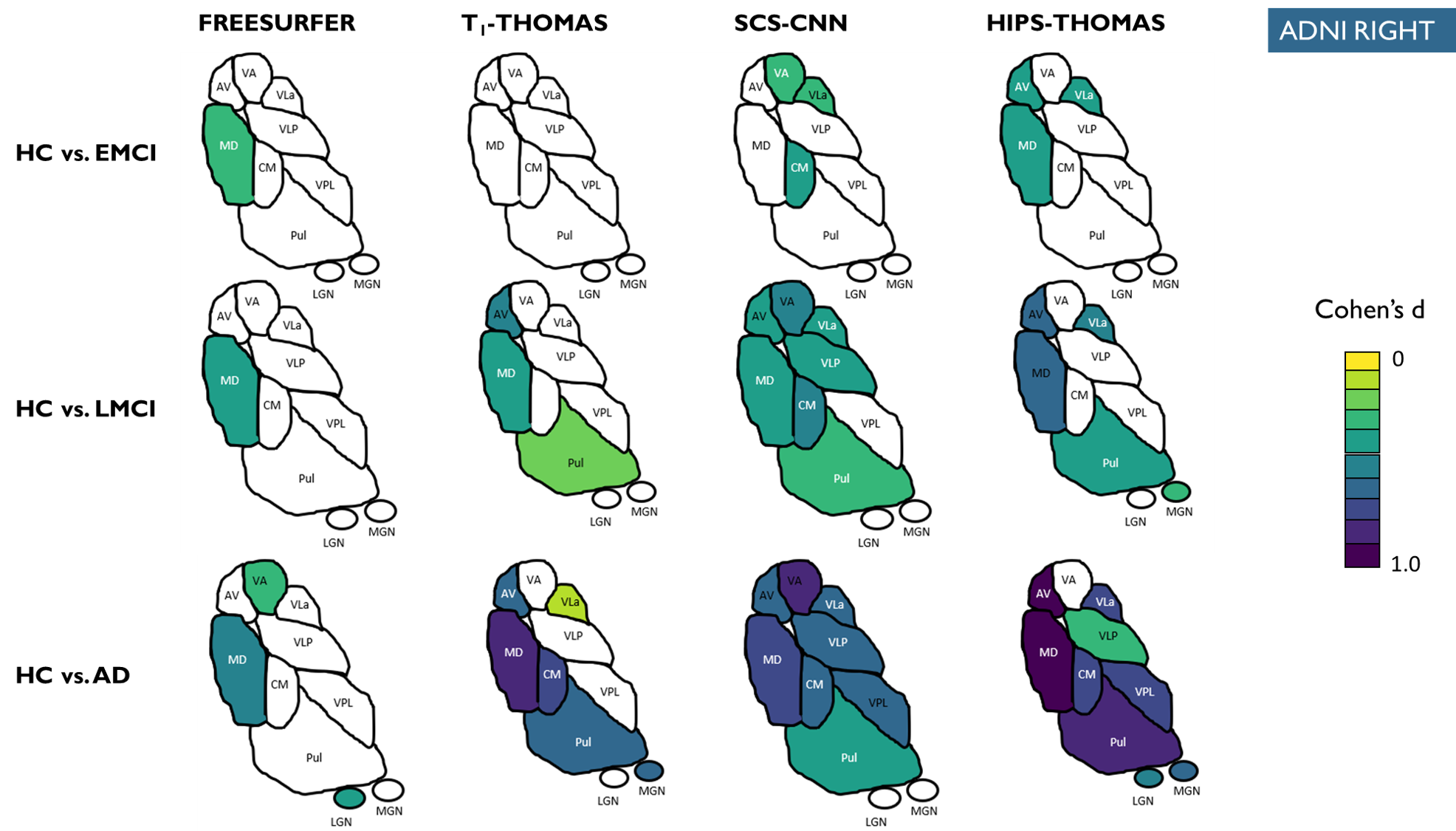
Supplementary figure 3:
Right hemisphere thalamic nuclei atrophy as a function of AD stage (i.e. EMCI, LMCI, AD) for the 4 methods colorized for statistically significant nuclei using effect size (Cohen’s d). Only nuclei with statistically significant differences in the ANCOVA tests are coloured.
References
1. J. E. Iglesias et al., ‘A probabilistic atlas of the human thalamic nuclei combining ex vivo MRI and histology’, NeuroImage, vol. 183, pp. 314–326, Dec. 2018, doi: 10.1016/j.neuroimage.2018.08.012.
2. A. S. Bernstein, S. Z. Rapcsak, M. Hornberger, M. Saranathan, and the A. D. N. Initiative, ‘Structural Changes in Thalamic Nuclei Across Prodromal and Clinical Alzheimer’s Disease’, J. Alzheimers Dis., vol. 82, no. 1, pp. 361–371, Jan. 2021, doi: 10.3233/JAD-201583.
3. L. Umapathy, M. B. Keerthivasan, N. M. Zahr, A. Bilgin, and M. Saranathan, ‘Convolutional Neural Network Based Frameworks for Fast Automatic Segmentation of Thalamic Nuclei from Native and Synthesized Contrast Structural MRI’, Neuroinformatics, vol. 20, no. 3, pp. 651–664, Jul. 2022, doi: 10.1007/s12021-021-09544-5.
4. J. P. Vidal et al., ‘Robust thalamic nuclei segmentation from T1-weighted MRI’. arXiv, Apr. 14, 2023. doi: 10.48550/arXiv.2304.07167.
5. B. Williams, E. Roesch, and A. Christakou, ‘Systematic validation of an automated thalamic parcellation technique using anatomical data at 3T.’, NeuroImage, vol. 258, p. 119340, Sep. 2022, doi: 10.1016/j.neuroimage.2022.119340.